The last decade brought changes to what could be called the Gold Standard for DNA/RNA sequencing. With more than a decade of experience in Next-Generation Sequencing, STAB VIDA offers the best possible value for your money regarding NGS technology. We have the capacity to perform your sequencing project with a high-level of confidence and always with personalised customer support.
Try our strategies to sequence your genomes, exomes, transcriptomes, target metagenomics, shotgun metagenomics. Apply massive parallel sequencing and enjoy the possibility of producing millions of sequencing reads, with or without bioinformatics analysis answering all your questions for a fair price
At STAB VIDA we offer a service of NGS, proposing different platforms: Illumina® Novaseq or MiSeq®, to deliver the best service for your needs. In addition, we also offer the possibility of bioinformatics analyses of your sequencing results.
If you need help designing your project, contact us by email (sales@stabvida.com) and our team of experts will help you define the best approach to reach your projects’ goal, adjusting the price to your needs. On the other hand, if you already have your project well defined and what you need is an official budget, log in to our website (Login) and request a quote in the NGS section, choosing the proper service option.
 You will obtain millions of sequences for each of your samples in a few days!
You will obtain millions of sequences for each of your samples in a few days!
METAGENOMICS/METABARCODING
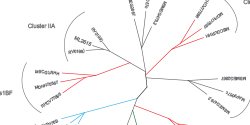
ITS and 16S rRNA target sequencing constitute well established amplicon sequencing methods used to identify and compare bacteria or fungi. These are proven methods for comparing sample phylogeny and taxonomy from complex microbiomes or environments. Additional targets, as CO1 can also be used for characterization of other species.
Our standard 16S protocol amplifies the V3-V4 region, using the following primer pair:
• 341F - CCTACGGGNGGCWGCAG
• 785R - GACTACHVGGGTATCTAATCC
Our standard ITS protocol amplifies the ITS1 region, using the following primer pair:
• ITS1f - CTTGGTCATTTAGAGGAAGTAA
• ITS2 - GCTGCGTTCTTCATCGATGC
Either using our standard targets or personalized ones, we can help you get the most information possible out of your samples. We might help you achieve this, through our QIIME2 based Metagenomics bioinformatics analysis service.
|
Contact us:
sales@stabvida.com
| tel: +351 210438606 | skype ID: Sales from STAB

RNA SEQUENCING
RNA-seq allows the detection and identification of nearly every class of molecules transcribed. Analyse whole transcriptomes in order to study gene expression analysis or marker discovery. RNA-seq doesn´t require prior knowledge of gene sequences. Get complete coverage of transcriptome and find information about exon structure and alternative splicing events.
mRNA-Seq
The mRNA-Seq library is based on an enrichment of polyadenylated mRNA. This. Poly(A) enrichment provides a high-sensitive option for detection of coding transcripts in samples without degradation.
Whole Transcriptome
The whole transcriptome library is based on rRNA depletion, enriching the sample for both mRNA and non-coding RNAs. This rRNA depletion strategy is great for samples that display a moderate degree of degradation.
Small RNA
Poly-A enrichment and complete transcriptome-based preparation methods do not allow an adequate evaluation of the Small RNAs, which require a specific sample preparation protocol, followed by an appropriate size selection. By following this specific protocol we can offer a complete service of discovery and profiling of Small RNAs.
The RNAseq data can then be serve multiple purposes, with our standard bioinformatics analysis service being suitable to characterize gene expression and differential gene expression.
Contact us:
sales@stabvida.com
| tel: +351 210438606 | skype ID: Sales from STAB

WHOLE GENOME SEQUENCING
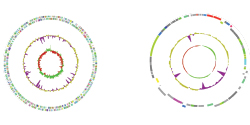
Single Species
Whole-genome sequencing (WGS) is a complete method for analysing entire genomes, providing the most comprehensive analysis of genome variance and structure. The WGS data can than be suitable for de novo assembly, annotation, or Mapping to reference & variant detection approaches, depending on the aim of the study.
Whole Genome Metagenomics (WGM)
Whole Genome Metagenomics (WGM), or shotgun metagenomic sequencing, is a powerful sequencing approach that provides insight into community biodiversity and function. Ultimately, the WMS seeks the sequencing all the genomes present in the samples. Dealing with all the generated information might be challenging, so you can also take advantage of our bioinformatics analysis service to help you get the most out of your data.
Lidar com toda a informação gerada pode ser um desafio, por isso também pode aproveitar o nosso serviço de análise de bioinformática para ajudá-lo a tirar o máximo partido dos seus dados.
Contacte-nos:
sales@stabvida.com
| tel: +351 210438606 | skype ID: Sales from STAB

Exome Sequencing

Whole Exome Sequencing (WES) intends to sequence the exonic regions of the genome. Despite constituting approximately only 1% of the human genome, most disease-related variants occur within this protein coding regions of the genome. With Exome sequencing you’ll be able to achieve high confidence variant calls and explore what they mean, either with comparison with appropriated databases or predictive analysis.
To achieve this, you can make use of our bioinformatics service, that goes from mapping to reference to the variant calling and variant annotation with multiple databases.
|
Contact us:
sales@stabvida.com
| tel: +351 210438606 | skype ID: Sales from STAB

Custom Projects

Run Only
Our Run Only Service is designed for customers who prefer to prepare their own libraries and thus have greater control of the entire process, or who want to prepare libraries using in-house workflows.
Epigenetics
DNA methylation is an important epigenetic mechanism capable of influencing the gene expression and structure of chromatin. The study of these mechanisms has particular relevance because methylation dysregulation is closely associated with multiple diseases. To enable this study, STABVIDA offers services of:
• Whole Genome Enzymatic Methylation Sequencing (WGEMS) – Allows you to detect cytosine methylation patterns throughout the genome, including cpG, CHH and CHG.
• Reduced Representation Bisulfite Sequencing (RRBS) - Consists of a way to sequence only CpG-rich regions of the genome through digestion with restriction enzymes.
Immunoprecipitate chromatin sequencing (ChIP-seq) is a method that allows the analysis of interactions between proteins and DNA. With our ChIP-seq service you can efficiently and accurately detect protein binding sites of interest, especially transcription factors and other chromatin-associated proteins.
|
Contact us:
sales@stabvida.com
| tel: +351 210438606 | skype ID: Sales from STAB

PLASMID AND MITOCHONDRIAL DNA SEQUENCING
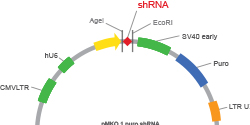
Get your plasmid constructs and your mtDNAs sequenced in 72h with 100x or 200x coverage.
Plasmids are essential tools used in genetic engineering for both gene transformation and genetic manipulation of prokaryotic and eukaryotic cells. They are very valuable when it comes to synthesizing proteins of interest in large quantities (e.g. insulin or antibiotics).
Mitochondrial DNA sequencing is a useful tool for studying certain cancers, aging, population genetics and biodiversity assessments. Also use our service to find out the heteroplasmic mutations from your samples
Using the Ion TorrentTM platform and expert bioinformatics software, we obtain high quality raw data and perform meticulous bioinformatics analysis, if desired. The raw data you get consists of up to 1 million reads of 200 bp each for the highest coverage of your DNA construct.
Contact us at: sales@stabvida.com | tel: +351 210438606 | skype ID: Sales from STAB
 You will obtain millions of sequences for each of your samples in a few days!
You will obtain millions of sequences for each of your samples in a few days!














 Français
Français  Español
Español  Português
Português  Nederlands
Nederlands  Deutsch
Deutsch